Improving COVID sequencing with DNA spike-ins
Our method to improve confidence and prevent common errors in SARS-CoV-2 sequencing is out in Nature Microbiology!
https://www.nature.com/articles/s41564-021-01019-2#Sec37
Sequencing the genomes from COVID19 is crucial to understand the spread and evolution of the virus. This may be the only paper I would have been happy to see become obsolete before publication, but alpha, beta, delta, and now omicron had other plans.
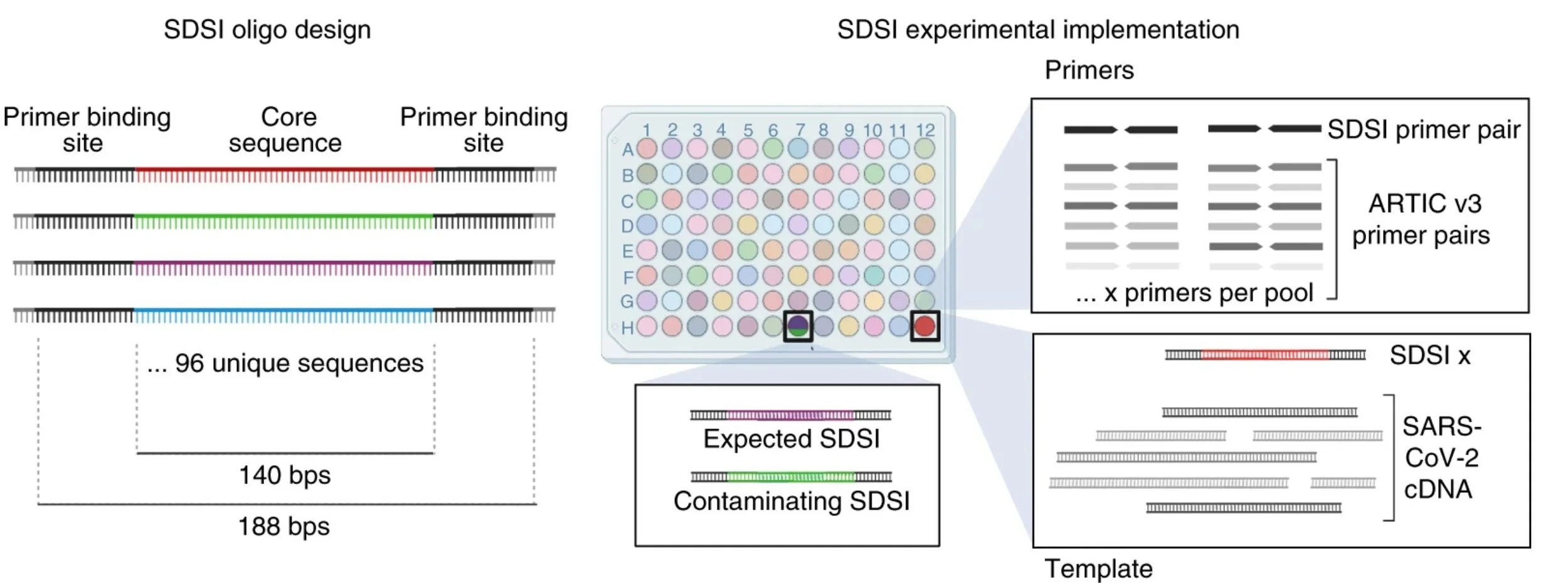
Amplicon methods can sequence small amounts of virus, but suffer from a high potential for contamination. We designed a set of 96 synthetic DNA spike-ins (SDSIs) to track individual genomes. We applied it to the Arctic Network’s deigns but its flexible to work on other primer sets or even other pathogens!
Our design uses a common primer binding site with unique core sequences. That means you add one extra primer set to the ARTIC mix, and an individual SDSI to each sample. The SDSIs get amplified along with the SARS-CoV-2 genome, tagging the sample.
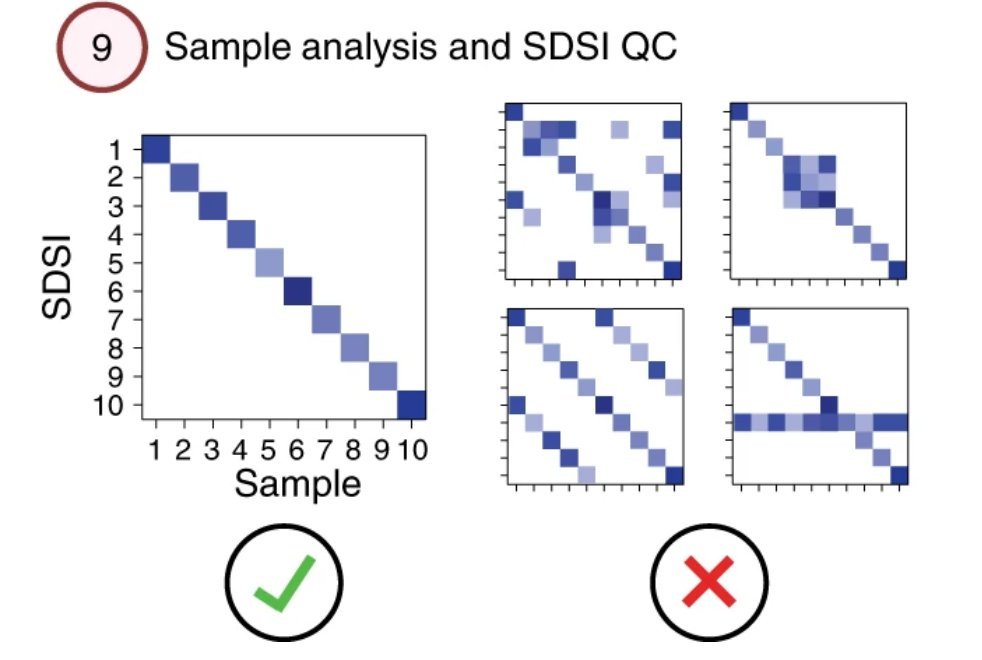
In sequencing the genome, you also sequence the SDSI. If you get the expected SDSI (and just the SDSI) great, no contamination or mixups! You'll get a plot like the left. But if you see unexpected SDSIs, it can flag these samples for further scrutiny (the plot on the right).
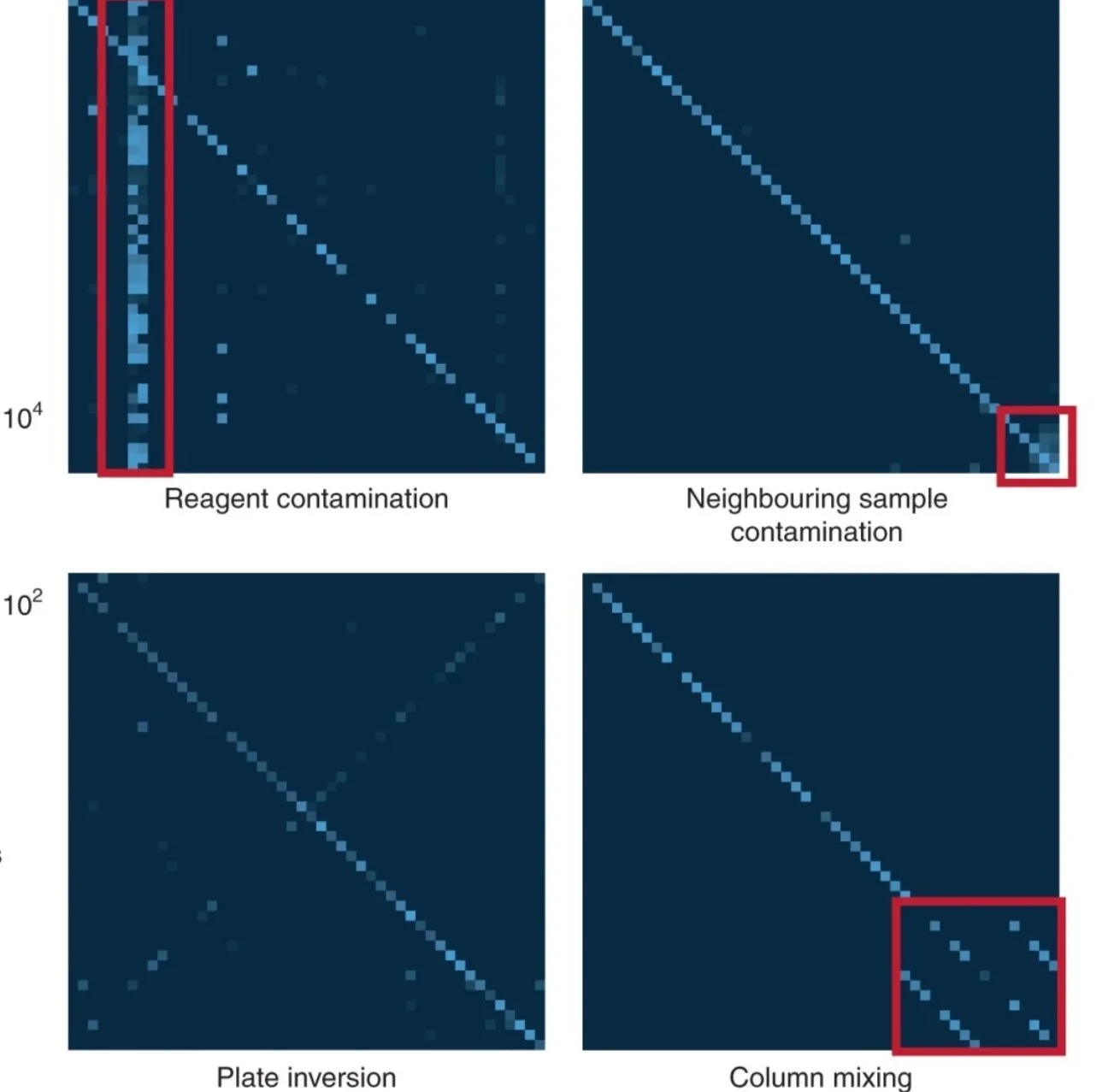
We've tested the method extensively in over 6000+ genomes (and counting). It works across viral loads and lineages. SDSIs detected all sorts or problems that would have be invisible with just water controls. Even with our experience processing thousands of samples, mistakes happen. The SDSIs detected reagent contamination, sample swaps, plate inversions, and more.
If undetected, these errors can skew epidemiological and clinical interpretations. In one case, we used SDSIs to detect a nosocomial transmission chain. As covid mutates slowly, SDSIs could confidently say identical genomes came from different people, not lab contamination.
We're continuing to improve this approach, including expanding designs for 384 spike-ins, application to other pathogens, and an RNA-spike in approach for end-to-end tracking. We're happy to share reagents, protocols, analysis pipelines, and expertise to other labs interested in applying this to their covid sequencing approaches. You can read over implementation for the ARTIC approach here: https://benchling.com/s/prt-R95g0tCxKOeCAqn8lAk3/edit. This work was done in an amazing collaboration between Katie Siddle and the Sabeti Lab!